 HOW DO BACTERIA CONTROL THE ATMOSPHERE?
HOW DO BACTERIA CONTROL THE ATMOSPHERE?
Microorganisms regulate atmospheric composition by consuming and producing various climate-relevant trace gases. For example, soil bacteria consume over 300 million tonnes of hydrogen and carbon monoxide from the atmosphere each year — they scavenge these gases using high-affinity metalloenzymes and use them as alternative energy sources for persistence. Other microorganisms regulate methane and carbon dioxide levels.
We have collected evidence that scavenging of atmospheric trace gases is widespread among microorganisms. We are now performing interdisciplinary studies to better understand the biochemical basis, physiological role, and ecological significance of trace gas scavenging and other gas-cycling processes. The knowledge is being used to help predict and mitigate greenhouse gas emissions.
Major projects
1. How do bacteria scavenge hydrogen and carbon monoxide? (ARC Discovery Project, 2020 – 2022)
2. Interactions between eutrophication and greenhouse gas production (ARC Discovery Project, 2021 – 2023)
3. Strategies to reduce methane emissions from livestock and agricultural soils (Global Research Alliance on Agricultural Greenhouse Gases)
4. Engineering bacteria to convert mixed waste gases into bioproducts (Monash Energy Institute Project)
Research team
Staff: Dr Sean Bay (postdoc), Dr Paul Cordero (postdoc), Ashleigh Kropp (research assistant)
Students: Bob Leung (PhD), David Gillett (PhD), Tess Hutchinson (PhD), Maha Alharbi (PhD), James Archer (Honours)
Collaborators: Prof Perran Cook, Dr Philipp Nauer, Dr Eleonora Chiri, A/Prof Matthew Stott, Dr Carlo Carere, Prof Holger Daims, Prof Rod Mackie, Dr Laura Meredith, Prof Damien Maher, Prof Scott Johnston, Prof Stefan Arndt, Dr Xiyang Dong, Prof Casey Hubert, Prof Graeme Attwood, Dr Min Wang
Previous findings
By pairing biogeochemical and microbiological analysis, we have revealed the dynamics of atmospheric methane cycling across diverse ecosystems, spanning ruminants, wetlands, oil seeps, termite mounds, and even trees (ISME J 2019, Nature Comms 2019, ISME J 2020a, Nature Comms 2021, New Phytologist 2021). Moreover, we have uncovered another phylum capable of mitigating methane emissions (Nature Microbiol 2021) and shown the methane-oxidising bacteria are a highly metabolically versatile group (ISME J 2016, ISME J 2017).
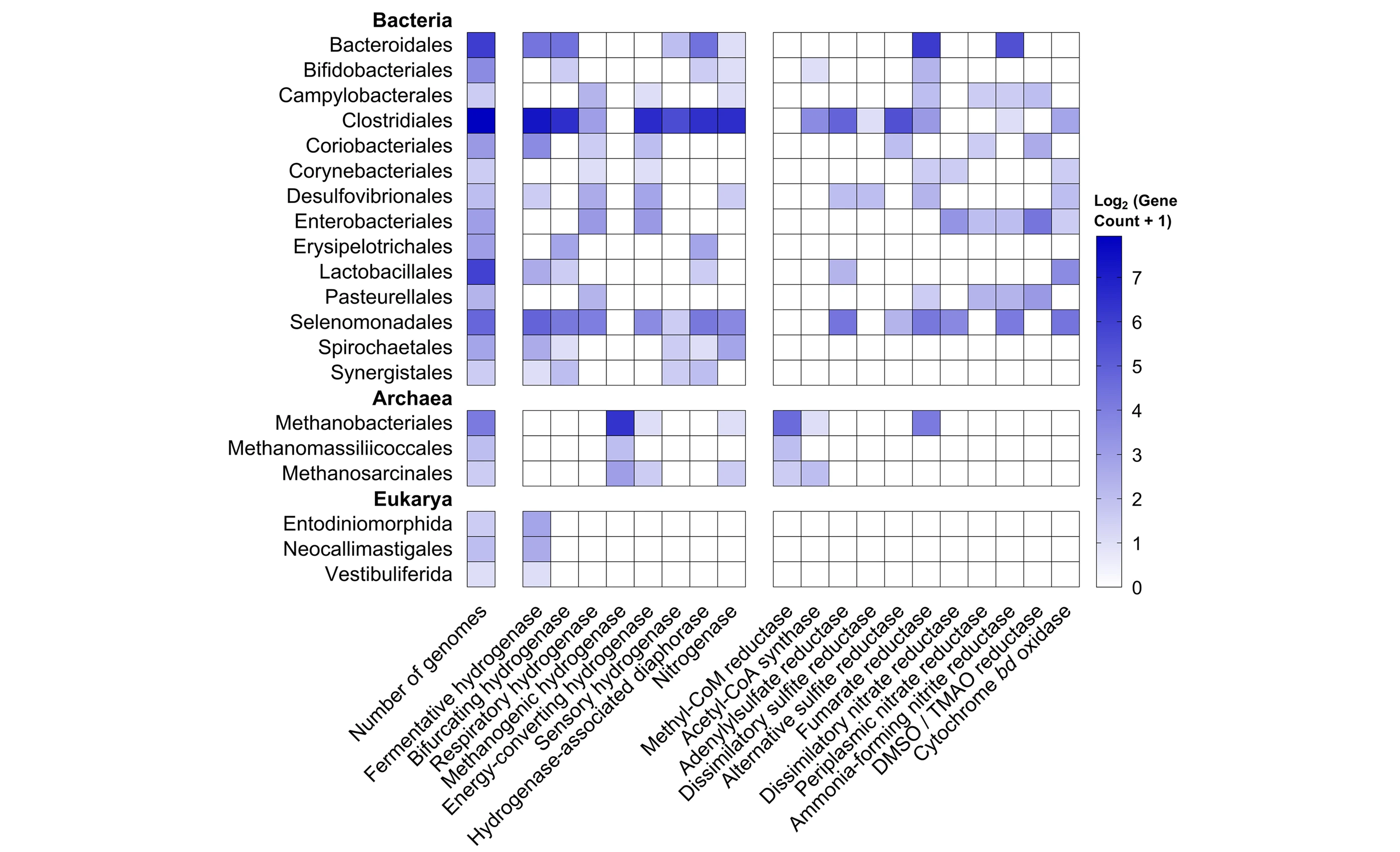
Figure: Example of our work on greenhouse gas metabolism. Genome survey revealing diverse pathways and organisms control the consumption and production of H2 in ruminants, and in turn influence substrate availability for methane-producing archaea (ISME J 2019).
More broadly, we are at forefront of resolving the mediators of the hydrogen and carbon monoxide cycles. We have shown that dormant soil microorganisms consume atmospheric hydrogen and carbon monoxide as dependable energy sources and resolved the enzymatic pathways involved (PNAS 2014a, PNAS 2014b, PNAS 2015, Nature 2017, ISME J 2019b, ISME J 2019c, ISME J 2020).